The Central England NERC Training Alliance (CENTA) has an open PhD position on tracking antimicrobial resistance across biomes, as part of the CENTA Flagship Project. To be hosted at the UK Centre for Ecology & Hydrology.
Overview
Globalization, growing and increasingly mobile human populations, intensive farming, chemical pollution, ecosystem degradation, and climate change have led to the emergence of new microbial pathogens and the spread of antimicrobial resistance (AMR). This raises concerns that host-environment connectivity in microbiome reservoirs may play a role in the ongoing AMR pandemic, contributing to more than a million deaths per year globally. The origins of AMR, however, lay in the complex interactions of microbial communities in natural environments. An often ignored, important aspect of AMR occurrence is its transport between biomes and hosts. AMR transport contributes to processes influencing microbial community assembly as well as being relevant in light of the One Health concept. Said concept was designed to counteract the global health challenges associated with the movement and evolution of AMR.
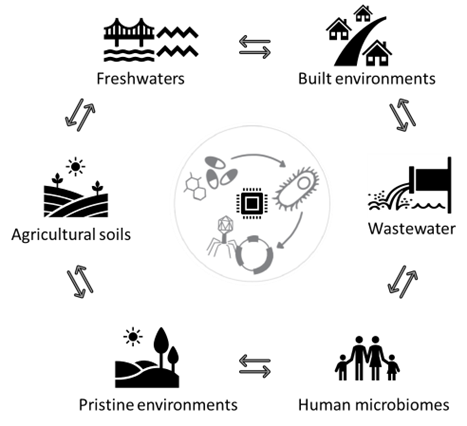
The Project
For this PhD studentship, you will characterise and track AMR genes across different natural and built environments. These include assessing AMR in the microbial fraction of soil (natural/pristine and agricultural), freshwater (rivers and lakes), wastewater and publicly available host-associated gut microbiome. The land-water interface is hypothesised to play an important role in the context of AMR transport, however, it has rarely been assessed. This project will employ molecular methods including metagenomic DNA sequencing, coupled with onsite/field sampling, wet- and dry-lab techniques. The goals of this research are three-fold as outlined below:
- Characterise patterns of AMR occurrence and diversity within pristine and anthropogenically influenced soils and freshwaters.
- Generate a metagenomically derived genome catalogue to explore the links between microbial taxa (MAGs) across biomes.
- Track the transport of AMR genes via MAGs at the land-water interface and vice versa.
Methodology
The project will comprise of a mix of fieldwork, wet- and dry-lab methodologies. The team will use data from national-scale and cross-biome surveys for this project. There will also be the opportunity to collect and analyse microbiomes from across wastewater, freshwater and soil interfaces. Using established protocols, you will extract DNA and generate metagenomic sequencing libraries, including long-read DNA sequencing, to characterize microbiomes. The team will process metagenomic data, from reads to the generation of MAGs, using existing and validated pipelines. Tools leveraging the updated Comprehensive Antibiotic Resistance Database (CARD) will provide AMR profiles of the MAGs. You will apply population genetics methodologies, including strain tracking based on single nucleotide polymorphisms, to understand AMR transport along the land-water interface and vice versa.
Further details
For any enquiries related to this project please contact Dr Susheel Bhanu Busi, UKCEH, susbus@ceh.ac.uk.
To apply to this project you can read the application process on the CENTA webpage. The application deadline is on Wednesday 8th January 2025.
