In this blogpost, Jochen Zubrod talks about developing an innovative machine learning approach for ecotoxicity predictions. These cutting-edge models show impressive predictive power for acute pesticide toxicity in freshwater organisms, holding promise for applications in environmental risk assessment and pesticide research and development.
Background
While the number of synthetic chemicals is ever-growing and demands increase to also understand potential environmental risks for non-standard species, it is both practically and legally impossible to test all species-chemical combinations of interest. Hence, predictive ecotoxicology using chemoinformatic techniques, such as quantitative structure-activity relationship (QSAR) models, received increasing attention in recent years. However, models presented so far are either restricted to a single species, preventing any cross-species predictions, or use taxonomy as a feature (i.e., independent variable) to this end. Although the latter allows to test new chemicals for all taxa the model was trained on, the categorial nature of this feature prevents any predictions for taxa beyond the training set. Hence, for models that are truly capable of both cross-chemical and cross-species predictions, surrogate numeric features are required that are readily available or easy to estimate for species of interest. For this purpose, variables describing species-specific physiological traits or processes – such as those defined in the context of Dynamic Energy Budget (DEB) theory – appear particularly well-suited as physiology has been shown to be an important driver of species sensitivity to chemicals.
Methodology
We pursued a multi-step machine learning (ML) modeling strategy combining data from multiple sources, while focusing our efforts to the prediction of acute pesticide toxicity in freshwater fish and invertebrates (i.e., LC50 and EC50 values). After identifying the optimal combinations of inter alia features and data balancing strategies for the Random Forest ML algorithm, we checked the suitability of our approach by feeding these optimal combinations to other popular or recently developed ML algorithms and manipulating our modeling pipeline. Finally, we applied a novel approach from the field of explainable (or interpretable) ML (i.e., SHAP) to investigate the importance of features to aid in creating a mechanistic understanding of chemical and physiological drivers of intrinsic sensitivity.
Results
Our efforts resulted in Bio-QSARs with R2 values on the independent test datasets of 0.85 and 0.83 for freshwater fish and invertebrates, respectively, and none of the alternative approaches could outperform these levels of predictive power. Moreover, as our datasets were pre-processed using an algorithmic approach for multi-collinearity correction, these models are fully explainable and methods to infer feature importance such as SHAP can be applied. SHAP analysis, in turn, showed that the involved DEB parameters were important for the models, particularly for the invertebrate Bio-QSAR (Fig. 1), which is likely related to the wider taxonomic spread of the invertebrate compared to the fish dataset.
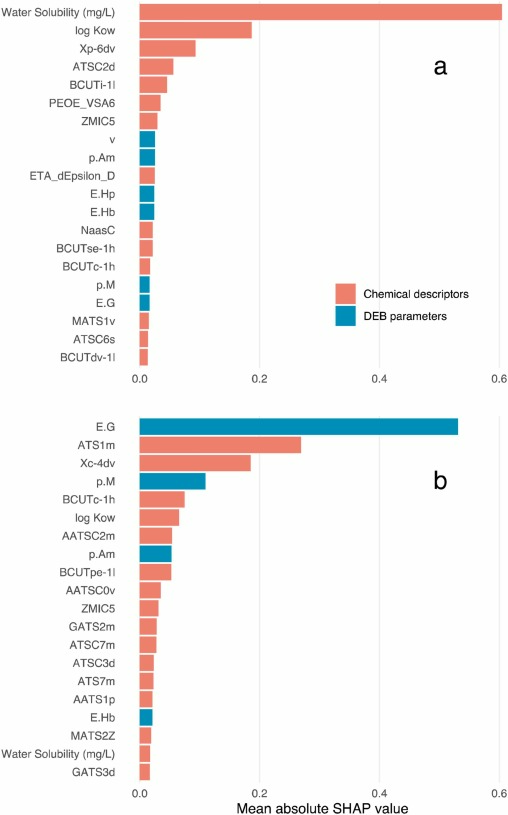
Conclusion
This study is the first to present a ML-based QSAR approach for (eco)toxicity predictions that is capable of flexible cross-chemical and cross-species predictions beyond levels present in training data. Moreover, predictive power of our Bio-QSARs compares favorably with existing models and tools for acute aquatic toxicity prediction. At the same time, our approach is applicable with a minimum of available information, namely the pesticide and the species whose combination is to be predicted – all required features can be either obtained from databases or predicted. This makes models based on our approach highly flexible and accessible for a number of applications related to ERA and pesticide research and development.
The paper titled ‘Physiological variables in machine learning QSARs allow for both cross-chemical and cross-species predictions’ was authored by Jochen P. Zubrod, Nika Galic, Maxime Vaugeois, and David A. Dreier and published open access in Ecotoxicology and Environmental Safety.
