In this new series of posts, research teams/groups of Landau’s Institute of Environmental Sciences introduce their Ecotoxicology-related research. This time: the group “Conservation Genetics”.
The team “Conservation Genetics”, led by Dr. Kathrin Theissinger, applies genetic and genomic methods as a tool to enable a better understanding and managing of natural populations, particularly with regard to biodiversity conservation and invasive species management. The team also addresses research questions concerning evolutionary ecotoxicology, i.e. the influence of pesticides in agricultural landscapes on the natural population structure.
The three main research topics currently are:
- Population genetics of Amphibians in Agricultural Landscapes
- Crayfish plague and Noble Crayfish Transcriptomics
- Chironomid Metabarcoding of temporary wetlands
Amphibians in Agricultural Landscapes

Photos by C. Brühl
In the DFG project Amphibians in agricultural landscapes: chemical landscape fragmentation implemented in a habitat modeling approach we are investigating the habitat use and population structure of amphibians in the monoculture vine between Landau/Pfalz and Neustadt/Weinstrasse.
We use a landscape genetics approach to assess the impact of chemical landscape fragmentation in viticulture for three amphibian species (Bufo bufo, Rana temporaria, Lissotriton helveticus) with varying dispersal abilities. We measure and compare gene flow rates and genetic diversities among breeding pond populations. The genetic data will be complemented with a telemetry study to investigate the migration direction from the aquatic to the terrestrial habitat. Additionally, we conduct interviews to management practices with local wine farmers to collect realistic land use data. Finally, we will implement the genetic and land use data in a habitat modeling approach that can be used to determine the population trend and improve the protection of amphibians in agricultural landscapes.
We are offering several student projects and theses. For students who are interested in participating in some part of this highly integrative project, please contact Christoph Leeb.
Crayfish plague and Noble Crayfish Transcriptomics

Photos by A. Schrimpf and D. Strand
In this field of research we are investigating the spread of the crayfish plague disease agent Aphanomyces astaci, the distribution of different genetic lineages of this oomycete pathogen, and the gene expression profiles of noble crayfish when infected with different virulent strains of this pathogen.
The noble crayfish (Astacus astacus) is a widely distributed decapod native species in European freshwaters. Its role in the aquatic ecosystem is paramount as it is one of the main keystone species in freshwater ecosystems and thus has significant impact on the biodiversity within its habitat. Today, all native European freshwater crayfish stocks are threatened by both the crayfish plague disease agent Aphanomyces astaci and invasive crayfish from North America, carrying and spreading this oomycete pathogen. Thus, also the noble crayfish is listed as vulnerable with a decreasing population status. We use an agent-specific real-time PCR analysis to detect the pathogen in wild populations and from eDNA samples. For determination of the different crayfish plague lineages we apply sequence and microsatellite analyses.
Invasive North American crayfish species are resistant against the crayfish plague infection as a consequence of coevolution. In European crayfish species, which are susceptible to the crayfish plague disease, the infection leads to crayfish death usually within a few days or weeks, depending on the pathogen strain and its virulence. In infection experiments of our finish colleagues (Jenny Makkonen, Harri Kokko and Japo Jussila from the University of Eastern Finland) significant differences were observed in the virulence of the A. astaci strains and in the resistance of different noble crayfish populations against the crayfish plague. Furthermore, also some natural populations of native European crayfish have been showing increased resistance, or at least prolonged carried status, towards acute crayfish plague infection. So what are the physiological reasons for this varied resistance among individuals of the same species? We aim to answers this question with next-generation-sequencing techniques. Using RNA-seq data we recently generated a de novo assembly and annotation of the noble crayfish transcriptome, which is a sound basis for further expression profile research. The analysis of such transcriptomic data is a key approach towards understanding how the crayfish plague infection affects the innate immune system of noble crayfish.
Chironomid Metabarcoding of Bti-treated temporary wetlands
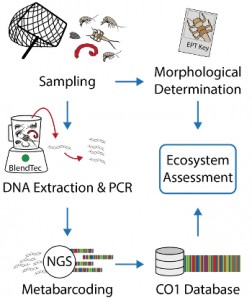
Figure by V. Elbrecht
In this study we test reliable next generation sequencing based metabarcoding tools to characterize the chironomid community compositions of temporal wetlands. This metabarcoding study is part of the MOSCOFFEE project led by Carsten Brühl.
As morphological species determination of some taxa is hardly possible for most people, DNA barcoding is a useful genetic alternative. Metabarcoding refers to determination of a community composition of a bulk sample by reading a specific barcoding region with next generation sequencing techniques. After DNA extraction from a bulk sample, which can contain hundreds or thousands of individuals from numerous taxa, a single step PCR with COI primers and subsequent sequencing in a high-throughput sequencing run is been conducted, resulting in millions of DNA sequences. Those barcodes then need to be bioinformatically processed and clustered into operational taxonomic units (OTUs), which then can be blasted in the Barcode of Life database (BOLD) for possible species IDs. For the metabarcoding methods and bioinformatics pipelines we collaborate with M.Sc. Vasco Elbrecht and Prof. Florian Leese from the University of Duisburg-Essen, who are leading researchers on DNA metabarcoding in Germany. By applying the above described metabarcoding tools on chironomid community samples of Bti-treated and –untreated wetlands we generated a neat dataset showing that the method works to reliably assess the chironomid diversity! Metabarcoding is thus a great alternative to classical morphological taxonomy to infer the chironomid community composition.
Team Members: Dr. Kathrin Theissinger, Dr. Anne Schrimpf, M.Sc. Jörn Panteleit, M.Sc. Christoph Leeb, Dipl.-Uwi Patrick Lenhardt, Dr. Jenny Makkonen (AufLand Fellow), B.Sc. Nina Keller, B.Sc. Elena Adams

You may also be interested in:
