In this post Anna Kästel and Kathrin Theissinger inform you what the new year brings: a new method for DNA based analyzing of environmental samples (metabarcoding), cooperations with the University of Duisburg- Essen and the University of Eastern Finland and hopefully exciting results about the sensitivity of chironomids to Bti!
In January 2016 we are going to analyze our first metabarcoding dataset. Metabarcoding is a DNA based technique to analyze environmental samples on species level. The target sequence (in our case COI) is amplified with next generation sequencing and compared to a database. The output of a sample (which can be thousands of different specimens in one tube!) is a list of all species in the sample. That makes metabarcoding to a prospering new tool to analyze biodiversity in a fast and reliable way. For the quite complicated bioinformatical analysis we need a little help from our friends Vasco Elbrecht from the University Duisburg-Essen and Jenny Makkonen from the University of Eastern Finland who will support us with their expertise.

The chironomids after drying process – hundreds of individuals, but how many species? (photo by A. Kästel)
The study background: In 2013 Anna Kästels Master thesis investigated the effect of Bti (Bacillus thuringiensis israelensis) based mosquito control on non-target chironomids. The Bti treatment showed statistically significant effects on the abundance of chironomids in these seasonal wetlands. Of special interest is not only the overall abundance but the species which showed a great sensitivity to Bti. With the help of metabarcoding we now want to find out the species composition in the study area. Particularly interesting are the species showing a high sensitivity to Bti treatment in early spring. These species could also have a higher impact on the foodweb since they are the only food resource for predators like newts or bats during the early spring.
How does the sample preparation work: In October 2015 Anna Kästel prepared chironomid samples gained in a field study in Geinsheim (near Neustadt/Weinstraße) in the year 2013. As first step the adult chironomids that were preserved in 70 % ethanol had to be dried and grounded to fine powder. As extraction method a bulk DNA extraction with a high salt extraction was conducted. After RNAse treatment and clean up the samples were ready for a travel to Essen.
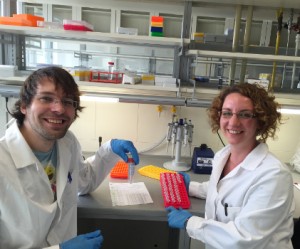
Vasco Elbrecht and Anna Kästel in the lab in Essen. Two weeks to prepare one small tube with all the DNA of about 2000 chironomid specimens! (photo by V. Elbrecht)
In Essen Vasco Elbrecht and Anna Kästel prepared the DNA library. In a first step the COI fragment was amplified. In the crucial PCR not only the fusion primers were inserted but also each sample was tagged individually. After the successful amplification the samples were purified and size selected with magnetic beads (SpriSelect). After all this steps the sample was ready for its trip abroad to Korea for next generation sequencing on a MiSeq.
While waiting for the data we got already trained how to operate the bioinformatic pipelines. Jenny Makkonen from Finland, Anna Kästel and Kathrin Theissinger joined the metabarcoding workshop in November 2015. Vasco Elbrecht and Florian Leese shared their metabarcoding expertise with us and 12 other participants. We got hands-on training with a high-throughput data set. See for further reading also the Leese Lab Blog.

